A rapid procedure to prepare for input file in a FASTA format for Alphafold3 local version
- 한국구조생물학회
- Biodesign
- Vol 13, No 1, Mar
-
2025.037 - 11 (5 pages)
-
DOI : 10.34184/kssb.2025.13.1.7
- 2
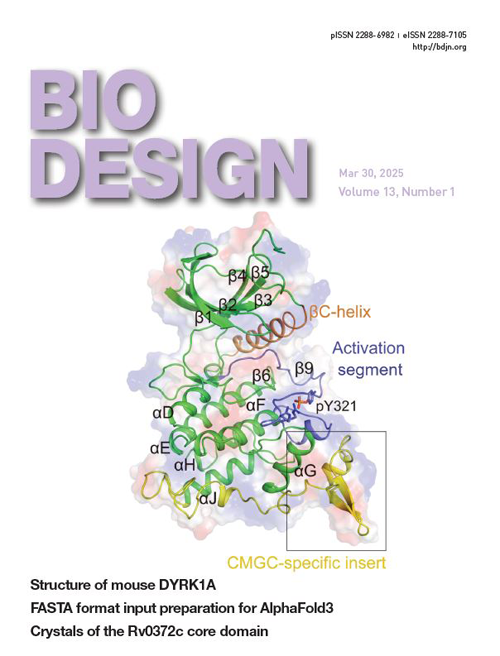
AlphaFold3 represents a significant advancement in protein structure prediction, extending its utility to complex biomolecular systems such as homomers, heteromers, nucleic acids, and ligand-bound structures. However, the local version of AlphaFold3 exclusively accepts JSON file inputs, which limits its practicality for simple structure predictions and high-throughput applications. This study developed a streamlined workflow utilizing FASTA format for input preparation, enhancing both the simplicity and efficiency of AlphaFold3 for local use, particularly in high-throughput structural predictions. Tools and scripts supporting this workflow are available at https://github.com/snufoodbiochem/Alphafold3_tools.
INTRODUCTION
RESULTS
DISCUSSION
ACKNOWLEDGEMENTS
CONFLICT OF INTEREST
REFERENCES
(0)
(0)